
A group of researchers harnessing the power of light to control gene expression has dramatically improved its method, optimizing speed and precision, and opening new research avenues for scientists who employ optogenetics — the use of light and genetic engineering to control cells.
A new paper in the journal Zebrafish details the advancements made in Professor Stephanie Woo’s lab and quantifies the results of experiments on zebrafish embryos.
Scientists use many tools to conduct genetic experiments, and Woo created TAEL, an inducible gene expression system, when she was a postdoctoral researcher at UCSF. TAEL uses an engineered transcription factor, a type of protein that can control gene expression. Unlike naturally occurring zebrafish transcription factors, TAEL was engineered to respond to light. By exposing zebrafish embryos to blue light, she and her colleagues can turn on expression of any gene of interest. Scientists can use this method to investigate gene function or to label cells with genetically encoded fluorescent proteins.
Since joining UC Merced in 2017, Woo, Professor Stefan Materna and colleagues have been working on TAEL 2.0. With help from lab manager Jesselynn LaBelle and others, they have not only improved the tool’s precision, they have also sped up the process.
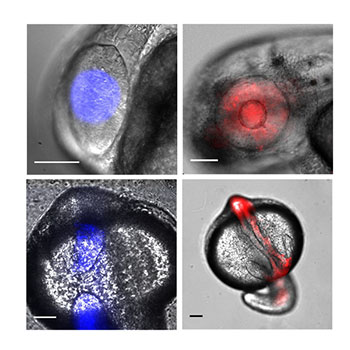
To optimize the tool for zebrafish, they added a nuclear localization signal (NLS) to the transcription factor to concentrate it in a cell’s nucleus, where the DNA is housed. They also changed the promoter — the DNA sequence that defines where the transcription factor will start gene transcription. These changes to both the transcription factor and the promoter were collectively referred to as TAEL 2.0.
“We study embryonic development of epithelial cells, so we’ll use TAEL 2.0 to get a better understanding of our genes of interest and their influence on that process,” explained Woo, a cell and developmental biologist. “But you could use this tool to look at any gene of interest — for example, one you suspect of causing cancer.”
As a test of TAEL 2.0’s capabilities, they used the system to drive expression of a red fluorescent protein called mCherry. When embryos are illuminated with blue light, the TAEL 2.0 system is activated and the embryos will glow red.
“TAEL 2.0 gives us tighter control of gene expression in both time and space,” Materna said. “This is faster than any chemical agent you could use, and if you want to express a gene in a specific area, all you do is turn on the light and the gene activates.”
With the change to the promoter, the researchers saw a 43.5 increase in the fold-induction rate of mCherry versus the old system where there was only a 2.9 fold-induction rate. By adding the NLS, they saw a 176.5 fold-induction rate of mCherry versus an 81.7 fold-induction rate with the old system, which lacked an NLS.
“It means we can activate more of the gene of interest, and we can do it faster, using less blue light,” said LaBelle, the paper’s lead author.
Woo said the TAEL tool doesn’t require expensive lab equipment, either.
“We use a $30 blue light panel we bought on Amazon, and we don’t use a fancy microscope,” she said. Zebrafish make good lab models because the eggs are externally laid and the embryos develop in less than one day.
“And they are clear, so we can see everything that’s going on inside,” she said.
Woo and Materna, a genetic engineer who worked on the regulatory element — the DNA that serves as the landing site for the transcription factor — are with the Department of Molecular and Cell Biology in the School of Natural Sciences. Both are affiliated with the Health Sciences Research Institute, and Woo is also part of the CREST Center for Cellular and Biomolecular Machines.
LaBelle, an undergraduate math major at California State University, Stanislaus, said her role in the development of the new version of TAEL was in data analysis.
“We could already see that a reporter gene we were looking at was brighter, but we needed the quantitative data to prove it,” she said.
LaBelle, a Dos Palos native, works full time in Woo’s lab and has another year of school to complete. She hopes to pursue her graduate degree at UC Merced.
“Before I started here, I knew very little about optogenetics,” she said. “Now, I am pretty well versed in it.”
Lorena Anderson

Senior Writer and Public Information Representative
Office: (209) 228-4406
Mobile: (209) 201-6255






